Use this to save results from a GLMM analysis in Genstat data structures.
- After selecting the appropriate boxes, type the identifiers of the data structures into the corresponding In: fields.
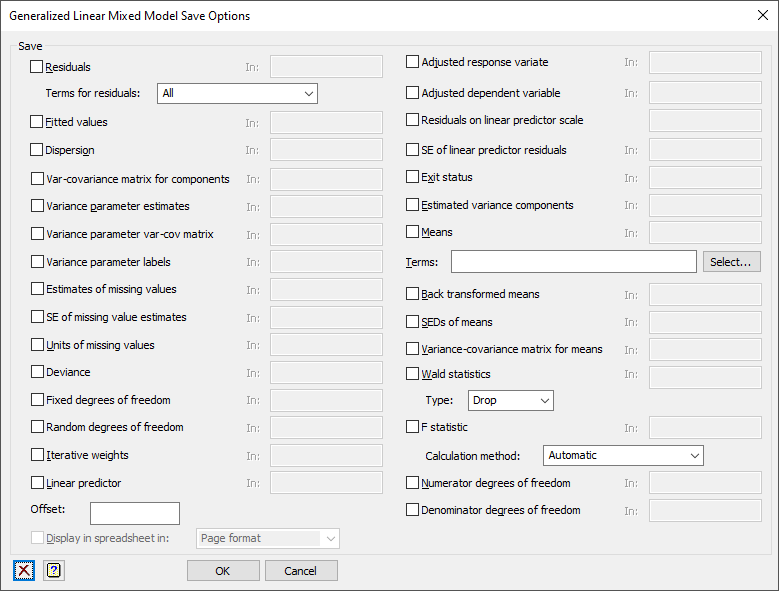
Save
Select the results you want to save.
| Residuals | Variate | Residuals from the fitted model using the specified Terms for residuals |
| Fitted values | Variate | Fitted values from the model |
| Dispersion | Scalar | The estimated dispersion |
| Components | Variate | Estimated variance components |
| Variance-covariance matrix for components | Symmetric matrix | Variance-covariance matrix of the variance components |
| Variance-covariance matrix for means | Symmetric matrix | Variance-covariance matrix of the means |
| Variance parameter var-cov matrix | Symmetric matrix | Variance-covariance matrix of the variance parameters |
| Variance parameter labels | Text | The descriptions of variance parameters |
| Estimates of missing values | Variate | The estimates for the missing values |
| SE of missing value estimates | Variate | The standard errors for the estimates for the missing values |
| Units of missing values | Variate | The unit numbers of the missing values. If you wanted to replace the missing values with their estimates, you would use CALC Y$[units] = mv_est |
| Deviance | Scalar | Deviance from the model – the type is specified by the Deviance to use option |
| Fixed degrees of freedom | Scalar | The number of degrees of freedom for the fixed model |
| Random degrees of freedom | Scalar | The number of degrees of freedom for the random model |
| Iterative weights | Variate | Iterative weights from the generalized linear model fitting |
| Linear predictor | Variate | Linear predictor from the generalized linear model fitting |
| Adjusted response variate | Variate | Adjusted response variate |
| Adjusted dependent variate | Variate | Adjusted dependent variate on the linear predictor scale |
| Residuals on linear predictor scale | Variate | Residuals from the fit on linear predictor scale |
| SE of linear predictor residuals | Variate | Standard errors of linear predictor residuals |
| Exit status | Scalar | The exit status from the GLMM procedure. A value of 0 indicates success. |
| Means | Table | Predicted means for the specified model terms |
| Back transformed means | Table | Back transformed means for the specified model terms |
| SEDS of means | Symmetric matrix | Standard errors of differences between means for the specified model term |
| Variance-covariance matrix for means | Symmetric matrix | Variance-covariance matrix for the means for the specified model term |
| F statistic | Scalar | The F statistic for the specified model term using the Calculation method |
| Numerator degrees of freedom | Scalar | The numerator degrees of freedom for the specified model term |
| Denominator degrees of freedom | Scalar | The denominator degrees of freedom for the specified model term |
Terms for residuals
The list allows selection from type of residuals that can be used.
| All | Use the residuals combined from all random terms. |
| Final | Use the residuals from the final random term. |
Wald statistics type
This option selects the type of Wald test used.
| Add | the tests are for adding terms sequentially to the model |
| Drop | the tests are for dropping terms from the full fixed model |
If Drop is selected, tests will only be made for terms which are not marginal to other terms in the model, e.g. for A + B + A.B, only the A.B term will be tested are A and B are marginal to A.B, and so cannot be dropped from the full model.
Terms for means and back transformed means
Specifies the model term for which tables of estimated means are to be saved. If tables of means are required for more than one model term, this menu should be invoked once for each term, changing the specification of the model term each time.
Calculation method for F-statistics
This controls whether Wald tests for fixed effects are accompanied with approximate F statistics and corresponding numbers of residual degrees of freedom. The computations, using the method devised by Kenward & Roger (1997), can be time consuming with large or complicated models. So, the default setting automatic, can be used to allow Genstat to assess the model itself and decide automatically whether to do the computations and which method to use. The other settings allow you to control what to do yourself:
| none | No F statistics are produced |
| algebraic | F statistics are calculated using algebraic derivatives (which may involve large matrix calculations) |
| numerical | F statistics are calculated using numerical derivatives (which require an extra evaluation of the mixed model equations for every variance parameter). |
Offset
The estimated means and back-transformed means will be estimated at the given offset. If this is not provided, the mean offset in the model will be used.
Display in spreadsheet
Select this to display the results in a new spreadsheet window. The format of the table of means and Back transformed means is controlled by the Format drop down list.
| Page format | Each table is displayed on a single page in a separate spreadsheet. The last specified classifying factor indexes the columns in the spreadsheet. |
| Column format | The tables occupy a single column but multiple tables can be put in a single spreadsheet. This is the default for a table with a single classifying factor. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Generalized Linear Mixed Model menu
- Generalized Linear Mixed Model Options dialog
- Generalized Linear Mixed Model Further Output dialog
- Generalized Linear Mixed Model Predictions dialog
- Generalized Linear Mixed Model Permutation Test dialog
- Generalized Linear Mixed Model Residual Plots dialog
- GLKEEP procedure
- GLMM procedure
- GLDISPLAY procedure
- GLPREDICT procedure
- GLPERMTEST procedure
- GLPLOT procedure