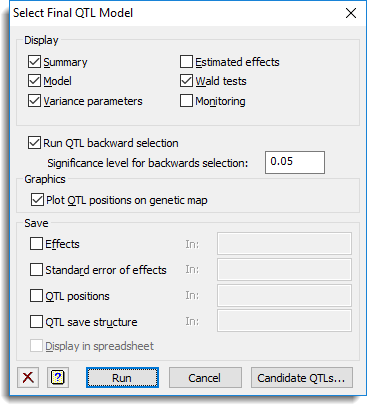
Use this to select and fit a final QTL model to estimate QTL effects for single environment trials. QTLs are selected from a list of candidate QTLs (loci) in single-environment trials by backward selection.
Display
This specifies which items of output are to be produced by the analysis.
| Summary | A summary of the QTLs retained in the model |
| Model | Description of the model fitted by the analysis |
| Variance parameters | Estimates of variance parameters |
| Estimated effects | Estimates of QTL effects |
| Wald test | Wald tests for all QTL effects in model |
| Monitoring | Monitoring output from the mixed model analysis |
Run QTL backward selection
When selected backward selection is used to determine a set of QTLs from the candidate QTLs.
Significance level for backward selection
The significance level to use at each step of the backward selection process.
Graphics
This specifies whether plots are to be produced by the analysis.
| Plot QTL positions on genetic map | Plots the significant QTLs on a genetic map. |
Save
These results can be saved when fitting the final QTL model. After selecting the appropriate boxes, you need to type the names for the identifiers of the data structures into the corresponding In: fields.
| Effects | Variate | Saves the estimated QTL effects. |
| Standard error of effects | Variate | Saves the standard errors of the QTL effects. |
| QTL positions | Variate | Saves the index numbers of the selected QTLs. This option is only enabled when the Run QTL backward selection option is selected. |
| QTL save structure | Pointer | Pointer to QTL save structure storing information and results for the significant effects. This structure cannot be displayed in a spreadsheet. |
Display in spreadsheet
Select this to display the results in a new spreadsheet window.
Action buttons
| Run | Select and fit the final QTL model. |
| Cancel | Close the dialog without further changes. |
| Candidate QTLs | Opens a dialog that lets you select or modify the set of candidate QTLs. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- QTL data space for using data in QTL menus
- QTL analysis using menus
- Single trait linkage analysis (single environment) menu
- Selecting candidate QTLs for final model dialog
- QSBACKSELECT and QSESTIMATE procedures for selecting a final QTL model for a single environment trial in command mode