Use this to select candidate QTLs when establishing a final QTL model to estimate QTL effects.
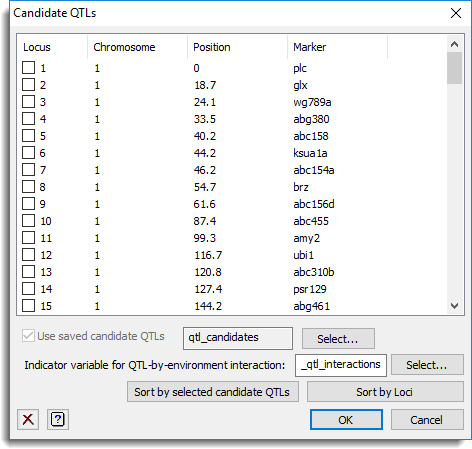
A list of the loci positions, chromosome groupings, positions within chromosome and virtual marker names are displayed. The list shows selected locus numbers that are the candidate QTLs. You can estimate the final model using the selected QTLs or select alternative locus numbers. Within the list selected candidate QTLs are indicated by a tick in the box for the locus number and will have the row coloured in grey. A previously saved set of candidate QTLs can be loaded using the use saved candidate QTLs option.
Use saved candidate QTLs
When selected, the list of candidate QTLs will be selected using the loci index numbers from the data structure name supplied in the space provided. The list of candidate QTLs should be supplied in a variate of loci index numbers where the index numbers are within the range of loci displayed in the list. By default this uses the name of the structure used on the main menu from which this dialog was opened (e.g. single trait linkage analysis menu), but alternative candidate QTL variates can be selected by clicking on the Select button.
Indicator variable for QTL-by-environment interaction
For a multi-environment analysis this can be used to specify QTL-by-environment interaction for selected candidate QTLs. If backward selection is being run before estimating the final model then this can be used to supply an identifier name to store logical values indicating whether each candidate QTL showed a significant (1) or non-significant (0) interaction. If backward selection is not being run then this must be supplied. The variate should contain 1 or 0 values to represent significant or non-significant interactions. The length of this variate should match the number and order of the selected candidate QTLs. A name of a variate can be entered in the space provided or can be selected by clicking on the Select button.
Action buttons
| Sort by selected candidate QTLs | Sorts the list in ascending order of selected QTLs followed by the unselected QTLs. |
| Scan with cofactors | Sorts the list in ascending order of the loci index. |
| OK | Save the selected candidate QTLs and close the dialog. |
| Cancel | Close the dialog without further changes. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- QTL data space for using data in QTL menus
- Single trait linkage analysis (single environment) menu
- Single trait linkage analysis (multiple environments) menu
- Multi-trait linkage analysis (single environment) menu
- Linkage analysis options dialog.
- QSQTLSCAN, QMTQTLSCAN and QMQTLSCAN procedures for QTL genome scans in single and multiple environments in command mode.
- QCANDIDATES procedure for selecting QTLs in command mode.