Use this to save results from an AMMI analysis in Genstat data structures.
- After selecting the appropriate boxes, type the names for the identifiers of the data structures into the corresponding In: fields.
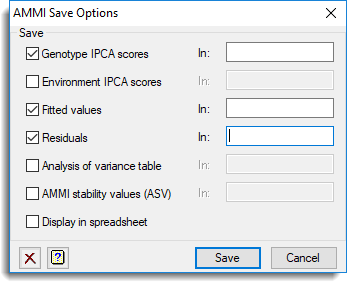
Save
| Genotype IPCA scores | Pointer | Containing a set of variates (each of length equal to the number of genotypes) to save the genotype IPCA scores |
| Environment IPCA scores | Pointer | To a set of variates to save the environment IPCA scores |
| Fitted values | Variate or Table | Saves the fitted values from the AMMI model |
| Residuals | Variate or Table | Saves the residuals from the AMMI model |
| Analysis of variance table | Pointer | Saves the analysis of variance table from the AMMI model |
| AMMI stability values (ASV) | Variate | Saves the stability values for each genotype from the AMMI model (the definition of ASV is given in the AMMI procedure help) |
Display in spreadsheet
The saved results will be displayed within a new spreadsheet window.