Use this to specify options for calculating the genetic predictors for a QTL analysis.
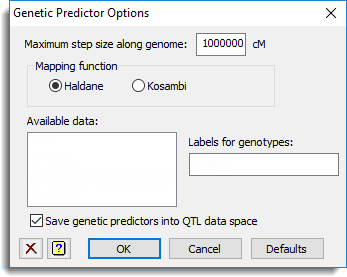
Maximum step size along genome
This specifies a maximum step size in cM to use for the calculation of the conditional probabilities. Setting this to a large value, such as the default value of 106, will calculate the IBD probabilities only at the marker positions.
Mapping function
This specifies the mapping function used. You can select either Haldane or Kosambi.
Available data
This lists text data structures currently available in Genstat appropriate for the labels for genotypes. Double-click a name to copy it to the current input field or type the name.
Labels for genotypes
A text specifying labels for the genotypes that correspond with the labels for the genotypes for the phenotypic means. This data structure can be used to ensure that there is correct matching for the genotypes between genotypic and phenotypic data. If a data structure exists in the QTL data space for the labels for genotypes then this will be automatically entered in the field.
Save genetic predictors into QTL data space
When the results are saved the data structure names will be added to the QTL data space so that the names of the structures will be present in the QTL menus.
Action buttons
| OK | Stores the option settings and closes the dialog. |
| Cancel | Close the dialog without making any changes. |
| Defaults | Resets the options to their default settings. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- QTL data space for using data in QTL menus
- Calculate genetic predictors menu.
- QIBDPROBABILITIES procedure for calculating genetic predictors in command mode.