Select menu: Stats | QTLs (linkage/Association) | Data Manipulation | Replace Missing Values in Marker Scores
Use this to replace missing marker scores with a score of the most similar genotype(s). It is possible that not all missing values will be replaced as the score of the most similar genotype(s) may also be a missing value.
- After you have imported your data, from the menu select
Stats | QTLs (linkage/Association) | Data Manipulation | Replace Missing Values in Marker Scores. - Fill in the fields as required then click Run.
You can set additional Options before running.
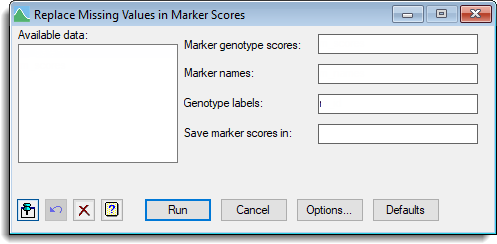
Available data
This lists data structures appropriate to the current input field. Double-click a name to copy it to the current input field or type the name. If data has been stored in a QTL data space then only the data structures present within that data space will be displayed in the Available data, otherwise all the current data within Genstat will be displayed. When data are present within the QTL data space you can right-click on the Available data list to open a shortcut menu where you can change between displaying data only within the data space and all data within Genstat.
Marker genotype scores
Specifies a pointer for the marker scores. The pointer should contain a set of factors (one for each marker) where each factor has the same labels representing the genotype scores and the labels are all in the same order.
Marker names
Specifies a text for the names of each marker.
Genotype labels
Specifies a text of the labels for the genotypes that correspond with the labels for the genotypes for the phenotypic means. This data structure is used to ensure that there is correct matching for the genotypes between genotypic and phenotypic data.
Save marker scores in
Specifies a pointer to save the marker scores that have had the missing scores replaced. The pointer contains a set of factors (one for each marker) where each factor has the same labels representing the genotype scores and the labels are all in the same order.
Action buttons
| Run | Run the analysis. |
| Cancel | Close the dialog without further changes. |
| Options | Opens a dialog where additional options and settings can be specified for the analysis. |
| Defaults | Reset options to the default settings. Clicking the right mouse on this button produces a pop-up menu where you can choose to set the menu using the currently stored defaults or the Genstat default settings. |
Action Icons
| Pin | Controls whether to keep the dialog open when you click Run. When the pin is down |
|
| Restore | Restore names into edit fields and default settings. | |
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Options for choosing which results to display
- QTL data space for using data in QTL menus
- QTL analysis using menus
- QMVREPLACE procedure in command mode