Use this to save results from calculating numbers of recombinations. These results are saved when the analysis is run and must be specified before running the analysis.
- After selecting the appropriate boxes, you need to type the names for the identifiers of the data structures into the corresponding In: fields.
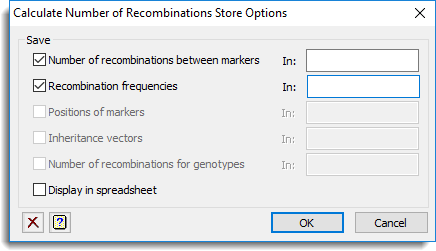
Save
The save options displayed will depend on the method selected on the Calculate number of recombinations menu. If a save option is not applicable for the chosen method it will be disabled. The available save options are as follows:
| Number of recombinations between markers | Symmetric matrix or pointer | For the two point method this saves the number of recombinations between markers. |
| Recombination frequencies | Symmetric matrix or pointer | For the two point method this saves the recombination frequencies. |
| Positions of markers | Variate | For the multi-point method this saves the positions of the markers. |
| Inheritance vectors | Pointer | For the multi-point method this saves inheritance vectors. |
| Number of recombinations for genotypes | Variate | For the multi-point method this saves the number of recombinations of the genotypes. |
Display in spreadsheet
Select this to display the results in a new spreadsheet window.
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Calculate number of recombinations menu.
- QRECOMBINATIONS procedure in command mode