Use this to specify options for the Cluster Probes/Genes menu.
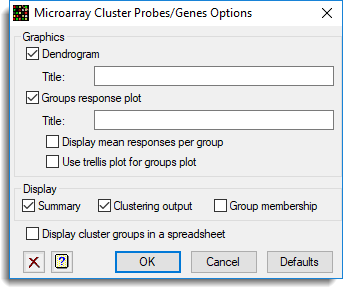
Dendrogram
When selected, produces a plot of the dendrogram. Use the Title field to specify a title for the plot. This option is only available when the Clustering method is set to Hierarchical.
Groups response plot
When selected, produces a plot of the profile of each group plotted against slide or target level. The y-axis will be the probe effect, and the x-axis will be the slide or target levels. If the Display mean response per group option is selected, then only the mean of the probes for each group for each slide/target will be plotted, otherwise each probe/gene will be plotted in a range of colours. Use the Title field to specify a title for the groups plot. By default each group is displayed within a separate plot, however, you can display the groups in different panels on a single graph by selecting the Use trellis plot for groups plot option. For a large number of probes, we recommend that the groups mean only be plotted when using the trellis plot option as this can be very memory intensive.
Display
This specifies which items of output are to be produced by the analysis.
| Summary | A summary of the data used to generate the similarity matrix |
| Clustering output | Displays output from the hierarchical or non-hierarchical clustering |
| Group membership | List of probes and targets that belong to each group generated by the respective cluster analyses |
Display cluster groups in a spreadsheet
Create a new spreadsheet containing the probe/gene names, group membership, and effects in columns, sorted by group. The effects columns will be labelled S[1],S[2], … where the first column corresponds to the effects for the first slide or target (as specified by the ordering of the targets/slides factor).
Action buttons
| OK | Save the options settings and close the dialog. |
| Cancel | Close the dialog without making any changes. |
| Defaults | Reset options to their default settings. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Cluster Probes/Genes menu
- Cluster Probes/Genes Store Options
- MAPCLUSTER procedure