Use this to form a pedigree matrix to take account of the parentage of the animals or genotypes. This dialog is activated by clicking the Generate button on the Individual/Animal Model dialog.
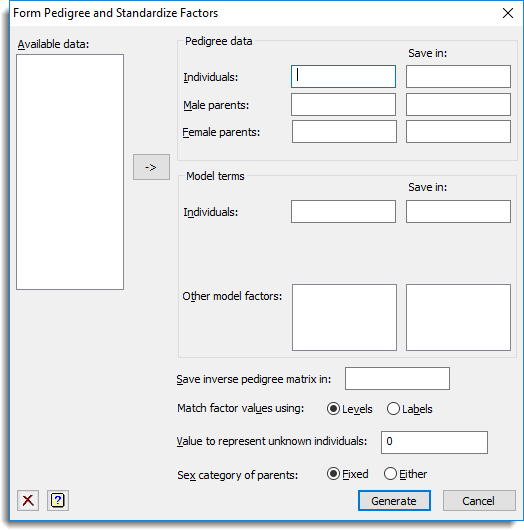
This pedigree information is specified by three factors: one that identifies the individuals for which data are available, and two others that indicate their male parents and their female parents (if available). The pedigree information is used to generate a sparse inverse relationship matrix that can then be used by the Individual/Animal Model or Parental Model menus to define a correlation model of the individual (or animal) effects in the analysis. To form a pedigree matrix the levels of the male and female factors must match with the individuals, these levels must be in ascending order, and the parents must be defined in the individuals factor before their offspring. To ensure that the pedigree factors have their levels in the correct order they are checked and pre-processed before the sparse inverse relationship matrix is formed.
Available data
This lists data structures appropriate to the current input field. Double-click a name to copy it to the current input field or type the name.
Pedigree data
This specifies the pedigree factors that are used to form the relationship matrix.
| Individuals | Individuals within the pedigree. The name of the factor to store the pre-processed individuals should be entered in the Save in field. |
| Male parents | Male parents (or dams) of the progeny. The name of the factor to store the pre-processed male parents should be entered in the Save in field. |
| Female parents | Female parents (or dams) of the progeny. The name of the factor to store the pre-processed female parents should be entered in the Save in field. |
Model terms
Specifies any other model terms involving the individuals in the pedigree to have there levels standardized with those within the pedigree.
| Individuals | Individuals on which data have been measured. The name of the factor to store the pre-processed individuals should be entered in the Save in field. |
| Other model factors | A list of other model factors that involve the individuals from the pedigree and require their levels standardized. The names of the factors to store the pre-processed male parents should be entered in the Save in field in parallel with the Other model factors list. |
Save inverse pedigree matrix
Specifies the name of a pointer to save the sparse form of the inverse pedigree matrix.
Match factor values using
Specifies whether the values in the pedigree factors are match using their levels or labels.
Value to represent unknown individuals
Missing values in any of the pedigree factors will be treated as coding for unknown individuals. This option allows you to specify an additional code to represent unknown individuals. This can be set to 0 or -1 if matching using levels, or ‘*’ or ‘0’ if matching using labels.
Sex category of parents
Specifies whether the individual can act as either a male or female parent. Alternatively, the setting fixed specifies that an individual can act as a male or female parent, but not both.
Action buttons
| Generate | Generate the pedigree matrix and close. |
| Cancel | Close the menu without further changes. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Individual/Animal models menu.
- Parental models menu.
- VFPEDIGREE procedure for pre-processing pedigree factors.
- VPEDIGREE directive for generating an inverse relationship matrix.