Use this to select phenotypic means data structures from a file to import for QTL analysis. This dialog opens when you select Trait means and click Open from the Open Phenotypic Data files dialog. When data are imported they are added to the QTL data space for use within the QTL menus.
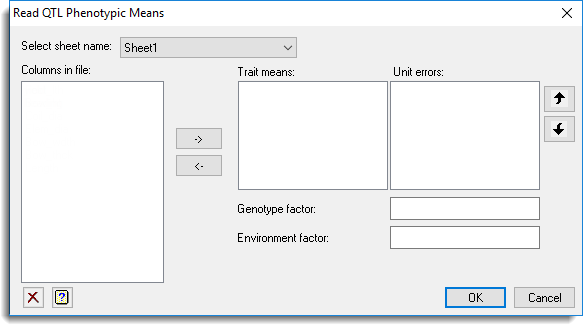
Select sheet name
If data are opened from a Genstat spreadsheet (.gsh or .gwb) file the sheets within the file will be listed. When a sheet name is selected the columns within that sheet will be displayed within the Columns in file option.
Columns in file
This lists data structures in the currently selected sheet for a Genstat spreadsheet file or the data structures within a file for other file types (i.e. comma delimited text files). Double-click a name or make a selection and click the ![]() to copy it to the current input field or type the name.
to copy it to the current input field or type the name.
Trait means
Specifies a list of variates that represent the means for different traits.
Unit errors
Specifies a list of variates containing the uncertainty on trait means (derived from individual unit or plot error) to be used in the QTL analysis. If the unit errors then they should be supplied for all the means in the Trait means list.
Genotype factor
Specifies a factor containing the genotypes. For comma delimited text files and MapQTL quantitative files a variate or text can be selected for the genotypes and will be automatically converted to a factor when the data are imported.
Environment factor
Specifies a factor specifying the different environments. For comma delimited text files and MapQTL quantitative files a variate or text can be selected for the environments and will be automatically converted to a factor when the data are imported.
Action buttons
| OK | Imports the data and stores the data structure names in the QTL data space. |
| Cancel | Close the dialog without making any changes. |
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Importing phenotypic data for QTL analysis
- QTL data space for using data in QTL menus