Use this to save results from two-way clustering. These results are saved when an analysis is run and must be specified before running an analysis.
- After selecting the appropriate boxes, type names for the data structures into the corresponding In fields.
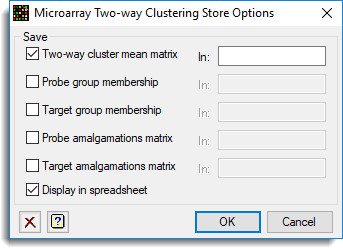
Save
The table below indicates the type of structures formed for each item.
| Two-way cluster mean matrix | Matrix | The tabulation of the Log-ratios by probe groups and target groups, as a two-way matrix. This contains data used by the shade plot. |
| Probe group membership | Factor | Stores the probe group membership for each probe or gene. The groups are specified by the Probe groups or Probe groups threshold % options for the hierarchical and non-hierarchical cluster methods respectively. |
| Target group membership | Factor | Stores the target group membership for each target or slide. The groups are specified by the Threshold % or Number of groups options for the Hierarchical and Non-hierarchical cluster methods respectively. |
| Probe amalgamations matrix | Matrix | Contains the probes or genes amalgamation data produced by hierarchical clustering. This is only available if the Cluster method is set to Hierarchical. |
| Target amalgamations matrix | Matrix | Contains the target or slides amalgamation data produced by hierarchical clustering. This is only available if the Cluster method is set to Hierarchical. |
Display in spreadsheet
Select this to display the results in a new spreadsheet window.
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |