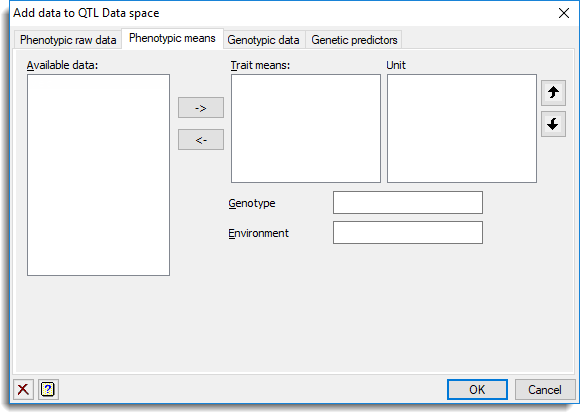
This can be used to add or remove phenotypic means and associated data structures in the QTL data space.
- To view this dialog click
 within the QTL Data view then click the Phenotypic means tab.
within the QTL Data view then click the Phenotypic means tab.

If data structure names have been stored in a QTL data space then when the QTL menus are opened the data structure names will automatically be entered into the relevant fields. Also only the data structures present within that data space will be displayed in the Available data, otherwise all the current data within Genstat will be displayed. When data are present within the QTL data space you can right-click on the Available data list to open a shortcut menu where you can change between displaying data only within the data space and all data within Genstat.
Available data
This lists data structures currently available in Genstat appropriate to the current input field. Double-click a name or by making a selection and clicking the ![]() button to copy the name to the current input field or type the name.
button to copy the name to the current input field or type the name.
Trait means
Specifies a list of variates that represent the means for different traits.
Unit
Specifies a list of variates containing the uncertainty on trait means (derived from individual unit or plot error) to be used in the QTL analysis. If the unit errors then they should be supplied for all the means in the Trait means list.
Genotype
Specifies a factor containing the genotypes.
Environment
Specifies a factor specifying the different environments for the trait means.
Action Icons
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- QTL data space for using data in QTL menus
- Adding phenotypic plot data, genotypic data or genetic predictor data to a QTL workbook.