Select menu: Stats | Microarrays | Cluster | Targets/Slides
Use this to cluster slides or targets together on the similarity of their responses over a number of probes or genes. The analysis uses hierarchical clustering using one of a range of link methods. The slides are grouped together so that the pattern of responses over the probes/genes are similar, with the groups as distinct as possible.
- After you have imported your data, from the menu select
Stats | Microarrays | Cluster | Targets/Slides. - Fill in the fields as required then click Run.
You can set additional Options before running the analysis and store the results by clicking Store.
A dendrogram for the hierarchical cluster analysis and a minimum spanning tree can be produced.
With large numbers of probes, many probes may be non-informative, only being subject to random variation, so an option of only using the top proportion of probes with largest mean absolute response is available.
Available data
This lists data structures appropriate for the field which currently has focus. Double-click a name to move it into the currently selected field or type the name in directly.
Data format
The data can be supplied in either of the following formats:
- Single variate for log-ratios with slide factor – All the log-ratios are stacked into a single variate, with factors that index the slide and probe/gene
- Pointer to log-ratio variates for each slide – Each slide has its data in a variate, and a pointer which points to this set of variates is provided. The Slides factor is not required, but if supplied it should just have one entry for each slide in the order of the variates in the pointer. The Probes/Genes factor is that for a single slide, and all slides must have a common layout.
The spreadsheet stack and unstack menus can be used to reorganise the data between these two formats.
Log-ratios
The log-ratios to cluster the slides on.
Targets or slides
The factor that identifies the slides. If the data are in pointer format, this has just one entry per slide, but if the data are in variate (stacked) format, this factor indexes the slides in the log-ratio variate.
Probes/Genes
The factor that identifies the probes or genes on a slide. If the data are in pointer format, this has just one entry per probe, but if the data are in variate (stacked) format, this factor indexes the probes in the log-ratio variate.
Link method
A number of methods for clustering are available and vary according to the way in which ‘closest’ is defined at each stage of merging groups. The following possibilities are available:
| Single link | Defines the similarity between two clusters as the maximum similarity between any two samples in those clusters |
| Nearest neighbour | Synonym for Single link |
| Complete link | Defines the similarity between two clusters as the minimum similarity between any two samples in those clusters |
| Furthest neighbour | Synonym for Complete Link |
| Average link | Defines the similarity between a cluster and two merging clusters as the average of the similarities with each of the original clusters. It therefore replaces two merging clusters by their mean, unweighted by cluster size |
| Group average | An average is taken over all the samples in the two merging clusters. Thus, the original clusters are replaced by their mean, weighted by cluster size |
| Median sorting | Can be thought of in terms of clusters being represented by points in a multidimensional space; when two clusters join, the new cluster is represented by the midpoint of the original cluster points |
Distance method
The method of combining the probe similarities together.
| Type | Contribution |
| Correlation | (1 + CORRELATION(Slide[i];Slide[j]))/2 |
| Euclidean Distance | SUM(1 – {(xi – xj) / range}**2) |
| City block Distance | SUM(1 – |xi – xj|/range) |
Use only top % of responding probes
Cluster using only the a percentage of the probes. These probes chosen will be those with largest average absolute responses.
Action buttons
| Run | Run the analysis. |
| Cancel | Close the dialog without further changes. |
| Options | Opens a dialog where additional options and settings can be specified for the analysis. |
| Defaults | Reset options to their default settings. Clicking the right mouse on this button produces a pop-up menu where you can choose to set the options using the currently stored defaults or the Genstat default settings. |
| Store | Opens a dialog to specify names of structures to store the results from the analysis. The names to save the structures must be supplied before running the analysis. |
Action Icons
| Pin | Controls whether to keep the dialog open when you click Run. When the pin is up |
|
| Restore | Restore names into edit fields and default settings. | |
| Graphics Output | Controls how graphs are to be drawn. You can either draw the graph in the Graphics View or save direct to files (JPEG, TIFF, EPS, EMF, GMF, BMP or PNG). | |
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
Example
The following dialog shows the clustering of a series of slides from a microarray experiment (Test_Slides.gsh):
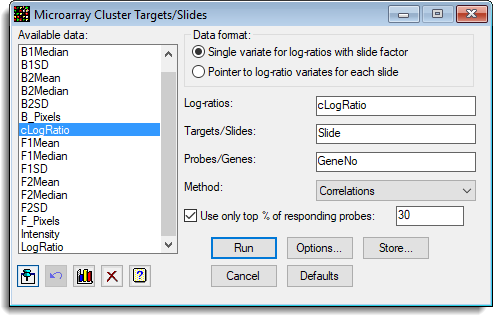
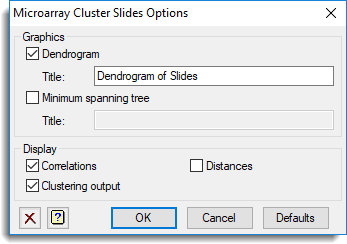
The options used were:
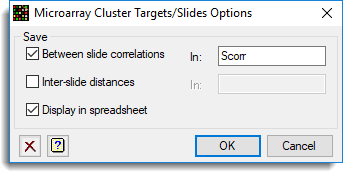
and the Store button as used to save Group results back to a spreadsheet:
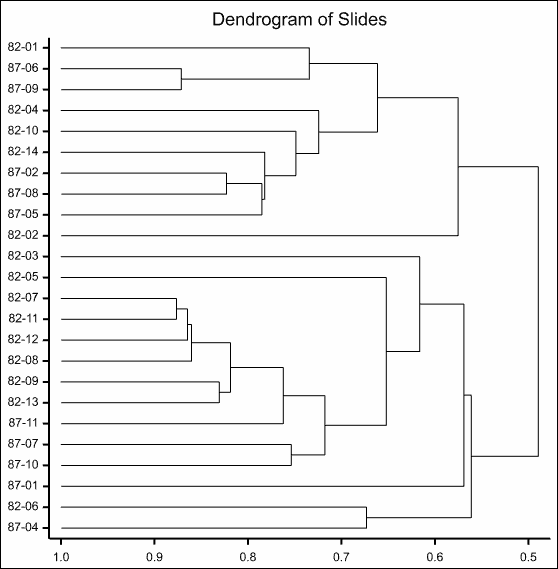
The resulting dendrogram is show below:
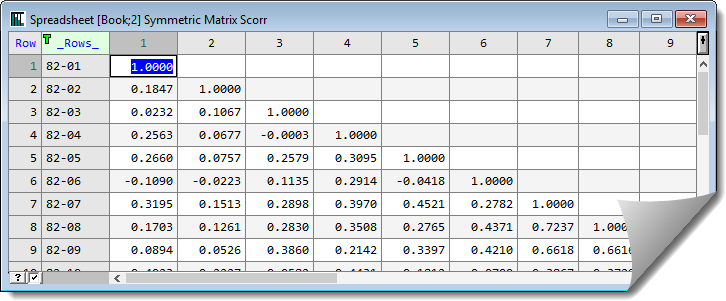
The saved correlation matrix will appear in a spreadsheet:
See also
- Cluster Targets/Slides Options
- Cluster Targets/Slides Store Options
- Cluster Probes/Genes
- Two-way Clustering
- Microarray Menus
- Two Channel Microarray Example
- Histograms, density plots, spatial plots and 2D plots for visualizing microarray data
- Volcano Plot
- Hierarchical Cluster Analysis menu
- Microarray procedures
- MASCLUSTER procedure
- HCLUSTER directive