Select menu: Stats | Regression Analysis | Mixed Models | Hierarchical Generalized Linear Models
This menu provides an interface to the facilities for fitting hierarchical generalized linear models (HGLMs) in the HG-system of John Nelder and Youngjo Lee.
- After you have imported your data, from the menu select
Stats | Regression Analysis | Mixed Models | Hierarchical Generalized Linear Models.
OR
Stats | Mixed Models (REML) | Hierarchical Generalized Linear Models. - Fill in the fields as required then click Run.
You can set additional Options then after running, you can save the results by clicking Save.
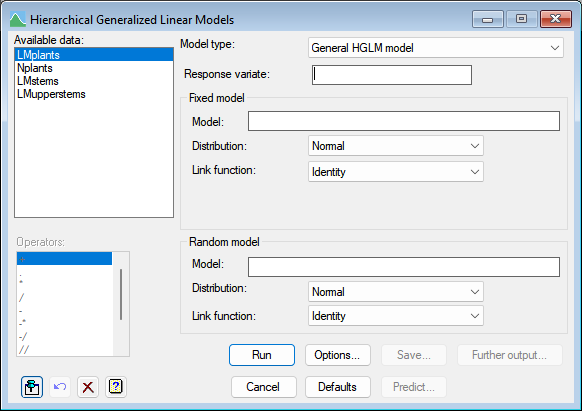
These models extend generalized linear models (GLMs) to include additional random terms in the linear predictor. They include generalized linear mixed models (GLMMs) as a special case, but do not constrain the additional terms to follow a Normal distribution and to have an identity link (as in the GLMM). For example, if the basic generalized linear model is a log-linear model (Poisson distribution and log link), a more appropriate assumption or the additional random terms might be a gamma distribution and a log link (for further details see the Methods section of the help information on HGANALYSE).
Available data
This lists data structures appropriate to the current input field. The contents will change as you move from one field to the next. Double-click on a name to copy it to the current input field or type the name.
Model type
This can be used to restrict the models (distributions and links) to specific model types.
| General HGLM model | Allow any selection of distributions and links for a general HGLM model |
| Conjugate HGLM model | Restrict random distribution and both links to give a conjugate HGLM model |
| GLMM model | Restrict random distribution and link to Normal and Identity to give a GLMM model |
Note: some distribution and link combinations are unlikely to produce a model that will converge or fit well.
The four conjugate HGLMs allowed are:
| Fixed distribution | Fixed link | Random distribution | Random link |
| Normal | Identity | Normal | Identity |
| Poisson | Logarithm | Gamma | Logarithm |
| Binomial | Logit | Beta | Logit |
| Gamma | Reciprocal | Inverse gamma | Reciprocal |
There is a second conjugate distribution for the Gamma distribution that uses a random Logarithm link instead of the Reciprocal link, but this needs to be specified as a General HGLM model.
Response variate
Specifies the response variate (dependent variate). This can be entered in directly or can be selected from those within the Available data list.
Fixed model
Specifies the fixed model terms for the HGLM.
Model
The terms to be fitted to the fixed model are specified by entering a model formula.
Distribution
List of available error distributions for the fixed model (this corresponds to error distribution of a GLM). If you select the binomial distribution, then you must supply the Binomial totals in the space provided.
Link function
Lists the available link functions for the fixed model.
Binomial totals
For a binomial distribution, this specifies a variate which contains the number of units the response was obtained from. The values in this must be greater than or equal to the corresponding values in the Response variate.
Censored data (for Poisson with log-link only)
When the option Censored are censored option is ticked, you can set a censoring value in upper bound (Direction: Right) or lower bound (Direction: Left) field at which values are censored. Right censoring means that you are certain the value is at least as great as the bound, but may be higher. This may occur when it is not possible to count above a particular value. Left censoring means that you are certain the value is at most as large as the bound, but may be lower. This may occur if you can not detect less than a given count.
Random model
Defines the random model for a hierarchical generalized linear model.
Model
The terms to be fitted to the random model are specified by entering a model formula.
Distribution
List of available error distributions for the random model.
Link function
Lists the available link functions for the fixed model.
Operators
This provides a quick way of entering operators in the fixed and random model formulas. Double-click on the required symbol to copy it to the current input field. You can also type in operators directly. See model formula for a description of each.
Options
This opens the Options dialog to set the options for the analysis.
Save
This opens the Save dialog to save results from the analysis.
Predict
This opens the Predictions dialog to form predictions and standard errors from the fitted model.
Further output
This opens the Further Output dialog to display more information, plots or a likelihood tests of fixed or random terms.
Action Icons
| Pin | Controls whether to keep the dialog open when you click Run. When the pin is down |
|
| Restore | Restore names into edit fields and default settings. | |
| Clear | Clear all fields and list boxes. | |
| Help | Open the Help topic for this dialog. |
See also
- Hierarchical Generalized Linear Model Options dialog
- HGLM Save Options dialog
- HGLM Further Output dialog
- HGLM Predictions dialog
- HGLM Likelihood Tests for Random Terms dialog
- HGLM Likelihood Tests for Fixed Terms dialog
- HGLM Model Checking for residual plots
- Fitted model menu for plotting the fit of an HGLM
- HGFIXEDMODEL, HGRANDOMMODEL, HGANALYSE and HGTOBITPOISSON procedures.
- HGRTEST procedure
- HGFTEST procedure
- HGNONLINEAR procedure